Coefficient field inversion in an elliptic partial differential equation
We consider the estimation of a coefficient in an elliptic partial differential equation as a first model problem. Depending on the interpretation of the unknowns and the type of measurements, this model problem arises, for instance, in inversion for groundwater flow or heat conductivity. It can also be interpreted as finding a membrane with a certain spatially varying stiffness. Let , be an open, bounded domain and consider the following problem:
where is the solution of
Here denotes the unknown coefficient field, the state variable, the (possibly noisy) data, a given volume force, and the regularization parameter.
The variational (or weak) form of the state equation:
Find such that
where is the space of functions vanishing on with square integrable derivatives.
Above, denotes the -inner product, i.e, for scalar functions defined on we write
and similarly for vector functions defined on we write
Gradient evaluation:
The Lagrangian functional is given by
Then the gradient of the cost functional with respect to the parameter is
where is the solution of the forward problem,
and is the solution of the adjoint problem,
Steepest descent method.
Written in abstract form, the steepest descent methods computes an update direction in the direction of the negative gradient defined as
where the evaluation of the gradient involve the solution and of the forward and adjoint problem (respectively) for .
Then we set , where the step length is chosen to guarantee sufficient descent.
Goals:
By the end of this notebook, you should be able to:
- solve the forward and adjoint Poisson equations
- understand the inverse method framework
- visualise and understand the results
- modify the problem and code
Mathematical tools used:
- Finite element method
- Derivation of gradient via the adjoint method
- Armijo line search
Import dependencies
from __future__ import print_function, division, absolute_import
import matplotlib.pyplot as plt
%matplotlib inline
import dolfin as dl
from hippylib import nb
import numpy as np
import logging
logging.getLogger('FFC').setLevel(logging.WARNING)
logging.getLogger('UFL').setLevel(logging.WARNING)
dl.set_log_active(False)
np.random.seed(seed=1)
Model set up:
As in the introduction, the first thing we need to do is to set up the numerical model.
In this cell, we set the mesh mesh, the finite element spaces Vm and Vu corresponding to the parameter space and state/adjoint space, respectively. In particular, we use linear finite elements for the parameter space, and quadratic elements for the state/adjoint space.
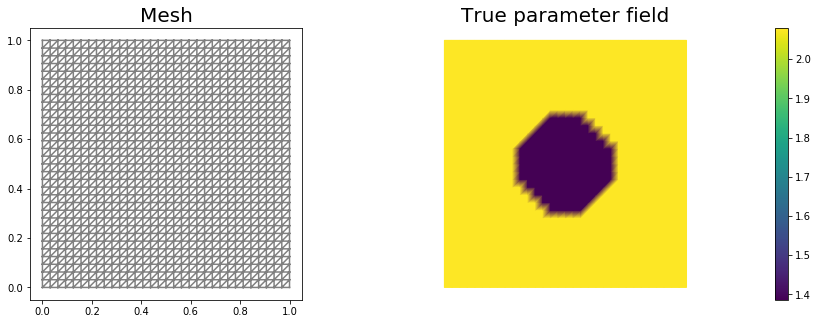
The true parameter mtrue is the finite element interpolant of the function
The forcing term f and the boundary conditions u0 for the forward problem are
# create mesh and define function spaces
nx = 32
ny = 32
mesh = dl.UnitSquareMesh(nx, ny)
Vm = dl.FunctionSpace(mesh, 'Lagrange', 1)
Vu = dl.FunctionSpace(mesh, 'Lagrange', 2)
# The true and initial guess for inverted parameter
mtrue = dl.interpolate(dl.Expression('std::log( 8. - 4.*(pow(x[0] - 0.5,2) + pow(x[1] - 0.5,2) < pow(0.2,2) ) )', degree=5), Vm)
# define function for state and adjoint
u = dl.Function(Vu)
m = dl.Function(Vm)
p = dl.Function(Vu)
# define Trial and Test Functions
u_trial, m_trial, p_trial = dl.TrialFunction(Vu), dl.TrialFunction(Vm), dl.TrialFunction(Vu)
u_test, m_test, p_test = dl.TestFunction(Vu), dl.TestFunction(Vm), dl.TestFunction(Vu)
# initialize input functions
f = dl.Constant(1.0)
u0 = dl.Constant(0.0)
# plot
plt.figure(figsize=(15,5))
nb.plot(mesh, subplot_loc=121, mytitle="Mesh", show_axis='on')
nb.plot(mtrue, subplot_loc=122, mytitle="True parameter field")
plt.show()
# set up dirichlet boundary conditions
def boundary(x,on_boundary):
return on_boundary
bc_state = dl.DirichletBC(Vu, u0, boundary)
bc_adj = dl.DirichletBC(Vu, dl.Constant(0.), boundary)
Set up synthetic observations:
To generate the synthetic observation we first solve the PDE for the state variable utrue corresponding to the true parameter mtrue.
Specifically, we solve the variational problem
Find such that
.
Then we perturb the true state variable and write the observation ud as
Here the standard variation is proportional to noise_level.
# noise level
noise_level = 0.01
# weak form for setting up the synthetic observations
a_true = dl.inner( dl.exp(mtrue) * dl.grad(u_trial), dl.grad(u_test)) * dl.dx
L_true = f * u_test * dl.dx
# solve the forward/state problem to generate synthetic observations
A_true, b_true = dl.assemble_system(a_true, L_true, bc_state)
utrue = dl.Function(Vu)
dl.solve(A_true, utrue.vector(), b_true)
ud = dl.Function(Vu)
ud.assign(utrue)
# perturb state solution and create synthetic measurements ud
# ud = u + ||u||/SNR * random.normal
MAX = ud.vector().norm("linf")
noise = dl.Vector()
A_true.init_vector(noise,1)
noise.set_local( noise_level * MAX * np.random.normal(0, 1, len(ud.vector().get_local())) )
bc_adj.apply(noise)
ud.vector().axpy(1., noise)
# plot
nb.multi1_plot([utrue, ud], ["State solution with mtrue", "Synthetic observations"])
plt.show()
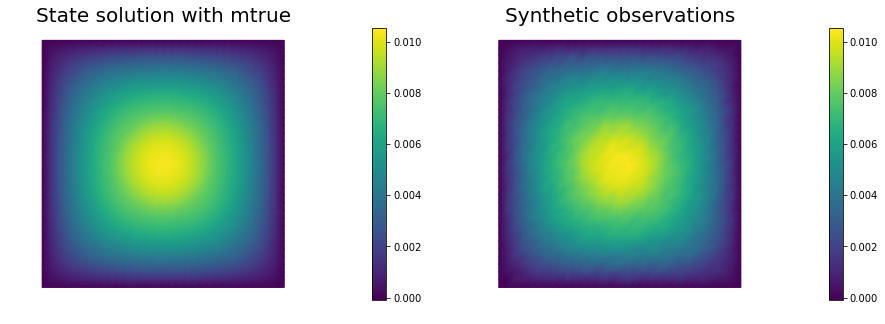
The cost functional evaluation:
# Regularization parameter
gamma = 1e-9
# Define cost function
def cost(u, ud, m, gamma):
reg = 0.5*gamma * dl.assemble( dl.inner(dl.grad(m), dl.grad(m))*dl.dx )
misfit = 0.5 * dl.assemble( (u-ud)**2*dl.dx)
return [reg + misfit, misfit, reg]
Setting up the variational form for the state/adjoint equations and gradient evaluation
Below we define the variational forms that appears in the the state/adjoint equations and gradient evaluations.
Specifically,
a_state,L_statestand for the bilinear and linear form of the state equation, repectively;a_adj,L_adjstand for the bilinear and linear form of the adjoint equation, repectively;CTvarf,gradRvarfstand for the contributions to the gradient coming from the PDE and the regularization, respectively.
We also build the mass matrix that is used to discretize the inner product.
# weak form for setting up the state equation
a_state = dl.inner( dl.exp(m) * dl.grad(u_trial), dl.grad(u_test)) * dl.dx
L_state = f * u_test * dl.dx
# weak form for setting up the adjoint equations
a_adj = dl.inner( dl.exp(m) * dl.grad(p_trial), dl.grad(p_test) ) * dl.dx
L_adj = - dl.inner(u - ud, p_test) * dl.dx
# weak form for gradient
CTvarf = dl.inner(dl.exp(m)*m_test*dl.grad(u), dl.grad(p)) * dl.dx
gradRvarf = dl.Constant(gamma)*dl.inner(dl.grad(m), dl.grad(m_test))*dl.dx
# Mass matrix in parameter space
Mvarf = dl.inner(m_trial, m_test) * dl.dx
M = dl.assemble(Mvarf)
Finite difference check of the gradient
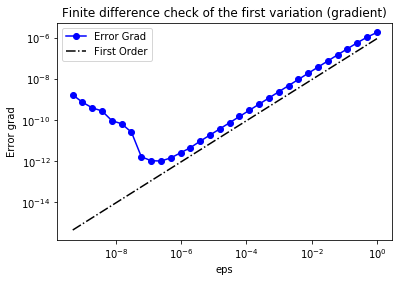
We use a finite difference check to verify that our gradient derivation is correct. Specifically, we consider a function and we verify that for an arbitrary direction we have
In the figure below we show in a loglog scale the value of as a function of . We observe that decays linearly for a wide range of values of , however we notice an increase in the error for extremely small values of due to numerical stability and finite precision arithmetic.
m0 = dl.interpolate(dl.Constant(np.log(4.) ), Vm )
n_eps = 32
eps = np.power(2., -np.arange(n_eps))
err_grad = np.zeros(n_eps)
m.assign(m0)
#Solve the fwd problem and evaluate the cost functional
A, state_b = dl.assemble_system (a_state, L_state, bc_state)
dl.solve(A, u.vector(), state_b)
c0, _, _ = cost(u, ud, m, gamma)
# Solve the adjoint problem and evaluate the gradient
adj_A, adjoint_RHS = dl.assemble_system(a_adj, L_adj, bc_adj)
dl.solve(adj_A, p.vector(), adjoint_RHS)
# evaluate the gradient
grad0 = dl.assemble(CTvarf + gradRvarf)
# Define an arbitrary direction m_hat to perform the check
mtilde = dl.Function(Vm).vector()
mtilde.set_local(np.random.randn(Vm.dim()))
mtilde.apply("")
mtilde_grad0 = grad0.inner(mtilde)
for i in range(n_eps):
m.assign(m0)
m.vector().axpy(eps[i], mtilde)
A, state_b = dl.assemble_system (a_state, L_state, bc_state)
dl.solve(A, u.vector(), state_b)
cplus, _, _ = cost(u, ud, m, gamma)
err_grad[i] = abs( (cplus - c0)/eps[i] - mtilde_grad0 )
plt.figure()
plt.loglog(eps, err_grad, "-ob", label="Error Grad")
plt.loglog(eps, (.5*err_grad[0]/eps[0])*eps, "-.k", label="First Order")
plt.title("Finite difference check of the first variation (gradient)")
plt.xlabel("eps")
plt.ylabel("Error grad")
plt.legend(loc = "upper left")
plt.show()
Initial guess
We solve the state equation and compute the cost functional for the initial guess of the parameter m0
m.assign(m0)
# solve state equation
A, state_b = dl.assemble_system (a_state, L_state, bc_state)
dl.solve(A, u.vector(), state_b)
# evaluate cost
[cost_old, misfit_old, reg_old] = cost(u, ud, m, gamma)
# plot
plt.figure(figsize=(15,5))
nb.plot(m,subplot_loc=121, mytitle="m0", vmin=mtrue.vector().min(), vmax=mtrue.vector().max())
nb.plot(u,subplot_loc=122, mytitle="u(m0)")
plt.show()
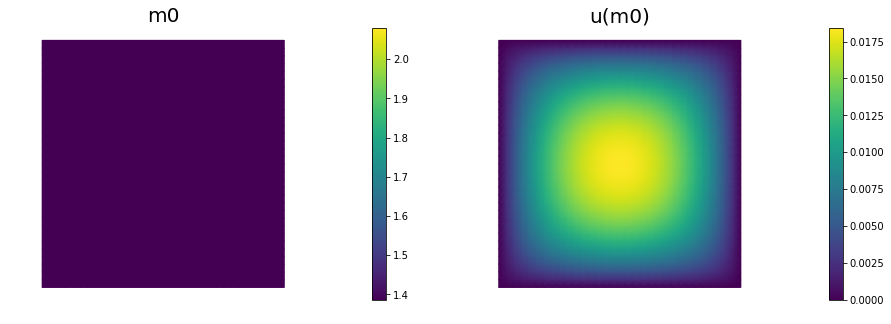
The steepest descent with Armijo line search:
We solve the constrained optimization problem using the steepest descent method with Armijo line search.
The stopping criterion is based on a relative reduction of the norm of the gradient (i.e. ).
The gradient is computed by solving the state and adjoint equation for the current parameter , and then substituing the current state , parameter and adjoint variables in the weak form expression of the gradient:
The Armijo line search uses backtracking to find such that a sufficient reduction in the cost functional is achieved. Specifically, we use backtracking to find such that:
# define parameters for the optimization
tol = 1e-4
maxiter = 1000
print_any = 10
plot_any = 50
c_armijo = 1e-5
# initialize iter counters
iter = 0
converged = False
# initializations
g = dl.Vector()
M.init_vector(g,0)
m_prev = dl.Function(Vm)
print( "Nit cost misfit reg ||grad|| alpha N backtrack" )
while iter < maxiter and not converged:
# solve the adoint problem
adj_A, adjoint_RHS = dl.assemble_system(a_adj, L_adj, bc_adj)
dl.solve(adj_A, p.vector(), adjoint_RHS)
# evaluate the gradient
MG = dl.assemble(CTvarf + gradRvarf)
dl.solve(M, g, MG)
# calculate the norm of the gradient
grad_norm2 = g.inner(MG)
gradnorm = np.sqrt(grad_norm2)
if iter == 0:
gradnorm0 = gradnorm
# linesearch
it_backtrack = 0
m_prev.assign(m)
alpha = 1.e5
backtrack_converged = False
for it_backtrack in range(20):
m.vector().axpy(-alpha, g )
# solve the state/forward problem
state_A, state_b = dl.assemble_system(a_state, L_state, bc_state)
dl.solve(state_A, u.vector(), state_b)
# evaluate cost
[cost_new, misfit_new, reg_new] = cost(u, ud, m, gamma)
# check if Armijo conditions are satisfied
if cost_new < cost_old - alpha * c_armijo * grad_norm2:
cost_old = cost_new
backtrack_converged = True
break
else:
alpha *= 0.5
m.assign(m_prev) # reset a
if backtrack_converged == False:
print( "Backtracking failed. A sufficient descent direction was not found" )
converged = False
break
sp = ""
if (iter % print_any)== 0 :
print( "%3d %1s %8.5e %1s %8.5e %1s %8.5e %1s %8.5e %1s %8.5e %1s %3d" % \
(iter, sp, cost_new, sp, misfit_new, sp, reg_new, sp, \
gradnorm, sp, alpha, sp, it_backtrack) )
if (iter % plot_any)== 0 :
nb.multi1_plot([m,u,p], ["m","u","p"], same_colorbar=False)
plt.show()
# check for convergence
if gradnorm < tol*gradnorm0 and iter > 0:
converged = True
print ("Steepest descent converged in ",iter," iterations")
iter += 1
if not converged:
print ( "Steepest descent did not converge in ", maxiter, " iterations")
Nit cost misfit reg ||grad|| alpha N backtrack
0 1.11658e-05 1.10372e-05 1.28638e-07 6.09224e-05 5.00000e+04 1
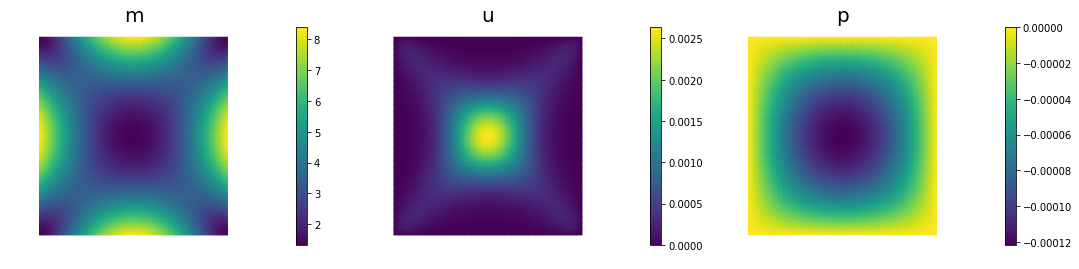
10 3.07468e-06 2.81411e-06 2.60567e-07 9.96798e-06 2.50000e+04 2
20 1.68333e-06 1.36733e-06 3.15999e-07 4.79045e-06 2.50000e+04 2
30 1.08642e-06 7.90406e-07 2.96014e-07 1.08801e-06 1.00000e+05 0
40 6.99077e-07 4.72703e-07 2.26374e-07 2.65053e-06 2.50000e+04 2
50 4.33194e-07 2.77234e-07 1.55960e-07 1.89540e-06 2.50000e+04 2
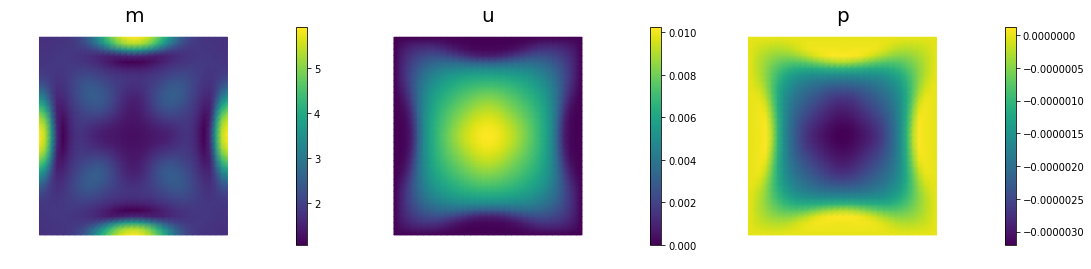
60 2.08625e-07 1.30743e-07 7.78817e-08 2.16983e-06 2.50000e+04 2
70 6.76060e-08 4.57894e-08 2.18166e-08 8.24665e-07 5.00000e+04 1
80 1.48917e-08 1.10392e-08 3.85253e-09 6.15229e-07 5.00000e+04 1
90 6.29351e-09 4.15392e-09 2.13959e-09 1.56127e-07 5.00000e+04 1
100 5.60737e-09 3.69792e-09 1.90946e-09 4.43594e-08 5.00000e+04 1
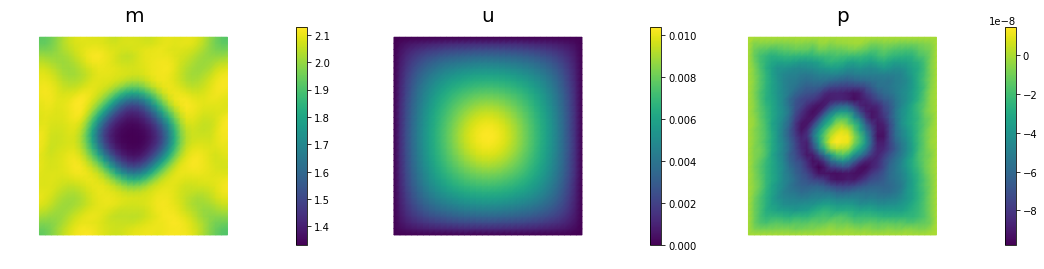
110 5.50313e-09 3.68332e-09 1.81981e-09 1.54409e-08 5.00000e+04 1
120 5.46016e-09 3.69724e-09 1.76293e-09 1.25966e-08 5.00000e+04 1
130 5.43430e-09 3.70647e-09 1.72783e-09 1.07081e-08 5.00000e+04 1
140 5.41730e-09 3.71189e-09 1.70542e-09 9.20452e-09 5.00000e+04 1
150 5.40583e-09 3.71517e-09 1.69066e-09 7.93538e-09 5.00000e+04 1
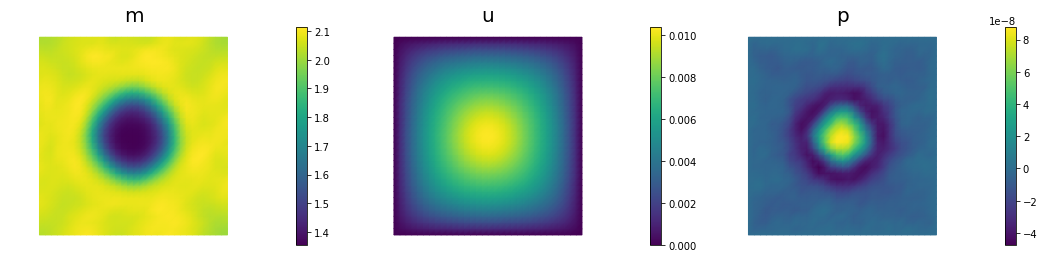
Steepest descent converged in 153 iterations
nb.multi1_plot([mtrue, m], ["mtrue", "m"])
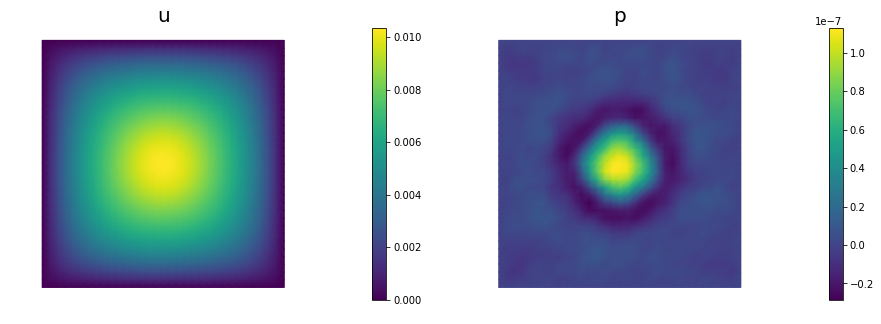
nb.multi1_plot([u,p], ["u","p"], same_colorbar=False)
plt.show()
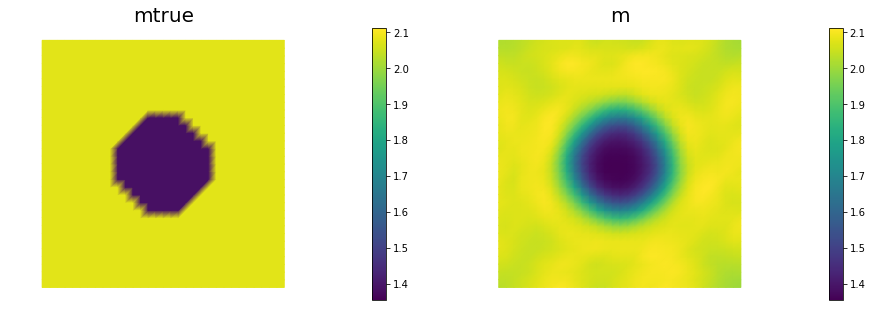
Hands on
Question 1
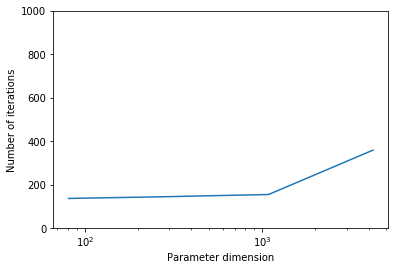
Report the number of steepest descent iterations for a discretization of the domain with , , , finite elements and give the number of unknowns used to discretize the log diffusivity field m for each of these meshes. Discuss how the number of iterations changes as the inversion parameter mesh is refined, i.e., as the parameter dimension increases. Is steepest descent method scalable with respect to the parameter dimension?
The number of interations increases as we refine the mesh. Steepest descent method is not scalable with respect to the parameter dimension.
Question 2
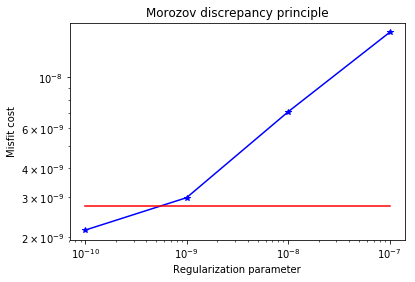
Add the advective term to the inverse problem and its hIPPYlib/FEniCS implementation and plot the resulting reconstruction of for a noise level of 0.01 and for a reasonably chosen regularization parameter.
See function def AddDiffInverseProblem(nx, ny, v, gamma, useTV, TVeps, plotting) for the implementation. Using Morozov’s discrepancy principle we chose the regularization parameter .
Question 3
Since the coefficient is discontinuous, a better choice of regularization is total variation rather than Tikhonov regularization, to prevent an overly smooth reconstruction. Modify the implementation and plot the result for a reasonably chosen regularization parameter . Use to make the non-differentiable TV regularization term differentiable ( has the same meaning as in Lab 2). In other words, your regularization functional should be:
See function def AddDiffInverseProblem(nx, ny, v, gamma, useTV, TVeps, plotting) for the implementation. Using Morozov’s discrepancy principle we choose the regularization parameter .
def AddDiffInverseProblem(nx, ny, v, gamma, useTV=False, TVeps=None, plotting=False):
np.random.seed(seed=1)
mesh = dl.UnitSquareMesh(nx, ny)
Vm = dl.FunctionSpace(mesh, 'Lagrange', 1)
Vu = dl.FunctionSpace(mesh, 'Lagrange', 2)
# The true and initial guess for inverted parameter
mtrue = dl.interpolate(
dl.Expression('std::log( 8. - 4.*(pow(x[0] - 0.5,2) + pow(x[1] - 0.5,2) < pow(0.2,2) ) )', degree=5),
Vm)
# define function for state and adjoint
u = dl.Function(Vu)
m = dl.Function(Vm)
p = dl.Function(Vu)
# define Trial and Test Functions
u_trial, m_trial, p_trial = dl.TrialFunction(Vu), dl.TrialFunction(Vm), dl.TrialFunction(Vu)
u_test, m_test, p_test = dl.TestFunction(Vu), dl.TestFunction(Vm), dl.TestFunction(Vu)
# initialize input functions
f = dl.Constant(1.0)
u0 = dl.Constant(0.0)
# set up dirichlet boundary conditions
def boundary(x,on_boundary):
return on_boundary
bc_state = dl.DirichletBC(Vu, u0, boundary)
bc_adj = dl.DirichletBC(Vu, dl.Constant(0.), boundary)
# noise level
noise_level = 0.01
# weak form for setting up the synthetic observations
a_true = dl.inner( dl.exp(mtrue) * dl.grad(u_trial), dl.grad(u_test)) * dl.dx \
+ dl.dot(v, dl.grad(u_trial))*u_test*dl.dx
L_true = f * u_test * dl.dx
# solve the forward/state problem to generate synthetic observations
A_true, b_true = dl.assemble_system(a_true, L_true, bc_state)
utrue = dl.Function(Vu)
dl.solve(A_true, utrue.vector(), b_true)
ud = dl.Function(Vu)
ud.assign(utrue)
# perturb state solution and create synthetic measurements ud
# ud = u + ||u||/SNR * random.normal
MAX = ud.vector().norm("linf")
noise = dl.Vector()
A_true.init_vector(noise,1)
noise.set_local( noise_level * MAX * np.random.normal(0, 1, len(ud.vector().get_local())) )
bc_adj.apply(noise)
ud.vector().axpy(1., noise)
if plotting:
nb.multi1_plot([utrue, ud], ["State solution with mtrue", "Synthetic observations"])
plt.show()
# Define cost function
def cost(u, ud, m, gamma):
if useTV:
reg = gamma * dl.assemble( dl.sqrt(dl.inner(dl.grad(m), dl.grad(m)) + TVeps)*dl.dx )
else:
reg = 0.5* gamma * dl.assemble( dl.inner(dl.grad(m), dl.grad(m))*dl.dx )
misfit = 0.5 * dl.assemble( (u-ud)**2*dl.dx)
return [reg + misfit, misfit, reg]
# weak form for setting up the state equation
a_state = dl.inner( dl.exp(m) * dl.grad(u_trial), dl.grad(u_test)) * dl.dx \
+ dl.dot(v, dl.grad(u_trial))*u_test*dl.dx
L_state = f * u_test * dl.dx
# weak form for setting up the adjoint equations
a_adj = dl.inner( dl.exp(m) * dl.grad(p_trial), dl.grad(p_test) ) * dl.dx \
+ dl.dot(v, dl.grad(p_test))*p_trial*dl.dx
L_adj = - dl.inner(u - ud, p_test) * dl.dx
# weak form for gradient
CTvarf = dl.inner(dl.exp(m)*m_test*dl.grad(u), dl.grad(p)) * dl.dx
if useTV:
gradRvarf = ( dl.Constant(gamma)/dl.sqrt(dl.inner(dl.grad(m), dl.grad(m)) + TVeps) )* \
dl.inner(dl.grad(m), dl.grad(m_test))*dl.dx
else:
gradRvarf = dl.Constant(gamma)*dl.inner(dl.grad(m), dl.grad(m_test))*dl.dx
# Mass matrix in parameter space
Mvarf = dl.inner(m_trial, m_test) * dl.dx
M = dl.assemble(Mvarf)
m0 = dl.interpolate(dl.Constant(np.log(4.) ), Vm )
m.assign(m0)
# solve state equation
A, state_b = dl.assemble_system (a_state, L_state, bc_state)
dl.solve(A, u.vector(), state_b)
# evaluate cost
[cost_old, misfit_old, reg_old] = cost(u, ud, m, gamma)
if plotting:
plt.figure(figsize=(15,5))
nb.plot(m,subplot_loc=121, mytitle="m0", vmin=mtrue.vector().min(), vmax=mtrue.vector().max())
nb.plot(u,subplot_loc=122, mytitle="u(m0)")
plt.show()
tol = 1e-4
maxiter = 1000
c_armijo = 1e-5
# initialize iter counters
iter = 0
converged = False
# initializations
g = dl.Vector()
M.init_vector(g,0)
m_prev = dl.Function(Vm)
while iter < maxiter and not converged:
# solve the adoint problem
adj_A, adjoint_RHS = dl.assemble_system(a_adj, L_adj, bc_adj)
dl.solve(adj_A, p.vector(), adjoint_RHS)
# evaluate the gradient
MG = dl.assemble(CTvarf + gradRvarf)
dl.solve(M, g, MG)
# calculate the norm of the gradient
grad_norm2 = g.inner(MG)
gradnorm = np.sqrt(grad_norm2)
if iter == 0:
gradnorm0 = gradnorm
# linesearch
it_backtrack = 0
m_prev.assign(m)
alpha = 1.e5
backtrack_converged = False
for it_backtrack in range(20):
m.vector().axpy(-alpha, g )
# solve the state/forward problem
state_A, state_b = dl.assemble_system(a_state, L_state, bc_state)
dl.solve(state_A, u.vector(), state_b)
# evaluate cost
[cost_new, misfit_new, reg_new] = cost(u, ud, m, gamma)
# check if Armijo conditions are satisfied
if cost_new < cost_old - alpha * c_armijo * grad_norm2:
cost_old = cost_new
backtrack_converged = True
break
else:
alpha *= 0.5
m.assign(m_prev) # reset a
if backtrack_converged == False:
print( "Backtracking failed. A sufficient descent direction was not found" )
converged = False
break
# check for convergence
if gradnorm < tol*gradnorm0 and iter > 0:
converged = True
print ("Steepest descent converged in ",iter," iterations")
iter += 1
if not converged:
print ( "Steepest descent did not converge in ", maxiter, " iterations")
if plotting:
nb.multi1_plot([mtrue, m], ["mtrue", "m"])
nb.multi1_plot([u,p], ["u","p"], same_colorbar=False)
plt.show()
Mstate = dl.assemble(u_trial*u_test*dl.dx)
noise_norm2 = noise.inner(Mstate*noise)
return Vm.dim(), iter, noise_norm2, cost_new, misfit_new, reg_new
## Question 1
ns = [8,16,32, 64]
niters = []
ndofs = []
for n in ns:
ndof, niter, _,_,_,_ = AddDiffInverseProblem(nx=n, ny=n, v=dl.Constant((0., 0.)),
gamma = 1e-9, useTV=False, plotting=False)
niters.append(niter)
ndofs.append(ndof)
plt.semilogx(ndofs, niters)
plt.ylim([0, 1000])
plt.xlabel("Parameter dimension")
plt.ylabel("Number of iterations")
Steepest descent converged in 135 iterations
Steepest descent converged in 143 iterations
Steepest descent converged in 153 iterations
Steepest descent converged in 357 iterations
Text(0,0.5,'Number of iterations')
## Question 2
n = 16
gammas = [1e-7, 1e-8, 1e-9, 1e-10]
misfits = []
for gamma in gammas:
ndof, niter, noise_norm2, cost,misfit,reg = AddDiffInverseProblem(nx=n, ny=n, v=dl.Constant((30., 0.)),
gamma = gamma, useTV=False, plotting=False)
misfits.append(misfit)
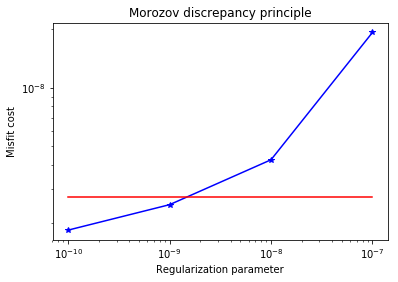
plt.loglog(gammas, misfits, "-*b", label="Misfit")
plt.loglog([gammas[0],gammas[-1]], [.5*noise_norm2, .5*noise_norm2], "-r", label="Squared norm noise")
plt.title("Morozov discrepancy principle")
plt.xlabel("Regularization parameter")
plt.ylabel("Misfit cost")
plt.show()
print("Solve for gamma = ", 1e-9)
_ = AddDiffInverseProblem(nx=n, ny=n, v=dl.Constant((30., 0.)), gamma = 1e-8, useTV=False, plotting=True)
Steepest descent converged in 233 iterations
Steepest descent converged in 154 iterations
Steepest descent converged in 139 iterations
Steepest descent converged in 256 iterations
Solve for gamma = 1e-09
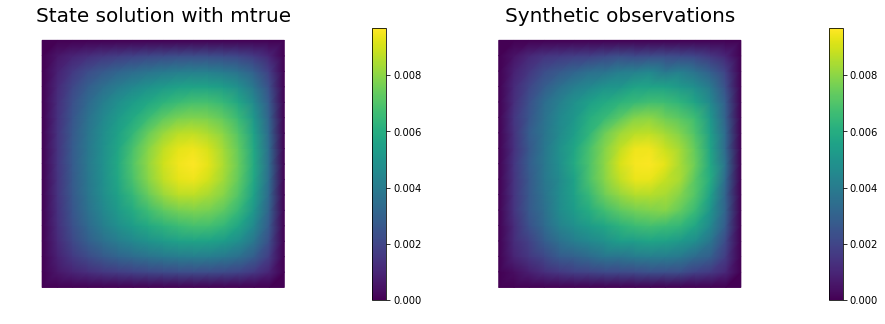
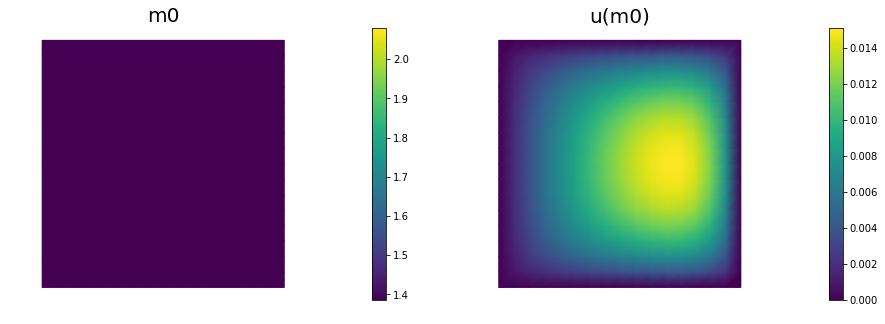
Steepest descent converged in 154 iterations
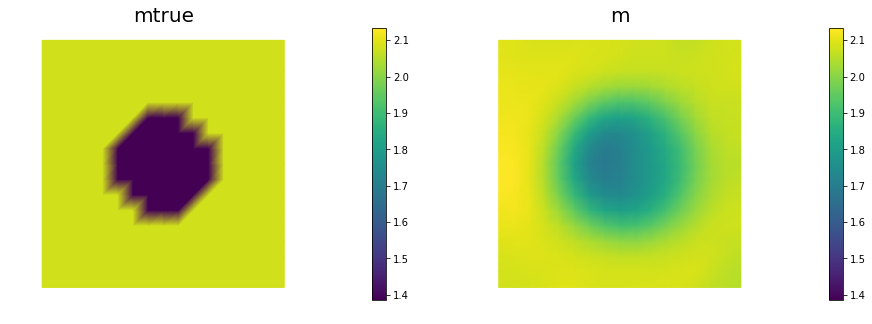
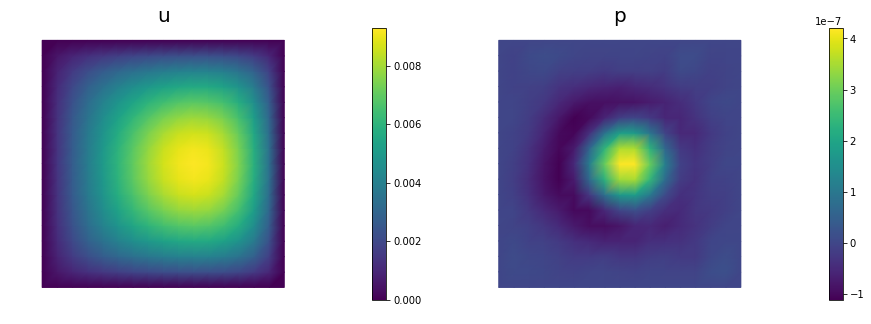
## Question 3
n = 16
gammas = [1e-7, 1e-8, 1e-9, 1e-10]
misfits = []
for gamma in gammas:
ndof, niter, noise_norm2, cost,misfit,reg = AddDiffInverseProblem(nx=n, ny=n, v=dl.Constant((30., 0.)),
gamma = gamma, useTV=True, TVeps=dl.Constant(0.1), plotting=False)
misfits.append(misfit)
plt.loglog(gammas, misfits, "-*b", label="Misfit")
plt.loglog([gammas[0],gammas[-1]], [.5*noise_norm2, .5*noise_norm2], "-r", label="Squared norm noise")
plt.title("Morozov discrepancy principle")
plt.xlabel("Regularization parameter")
plt.ylabel("Misfit cost")
plt.show()
print("Solve for gamma = ", 1e-8)
_ = AddDiffInverseProblem(nx=n, ny=n, v=dl.Constant((30., 0.)), gamma = 1e-8, useTV=True,
TVeps=dl.Constant(0.1), plotting=True)
Steepest descent converged in 928 iterations
Steepest descent converged in 376 iterations
Steepest descent converged in 192 iterations
Steepest descent converged in 267 iterations
Solve for gamma = 1e-08
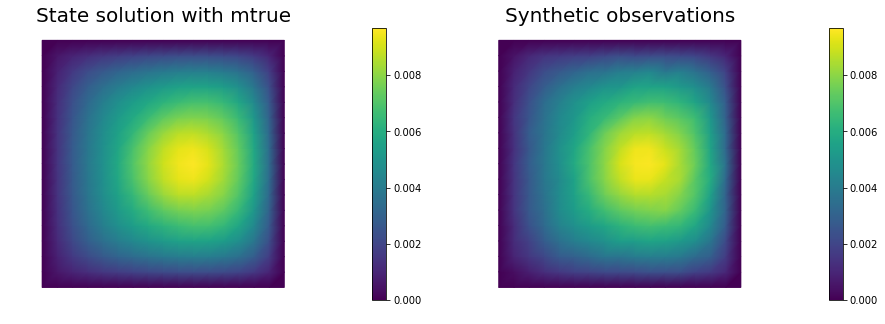
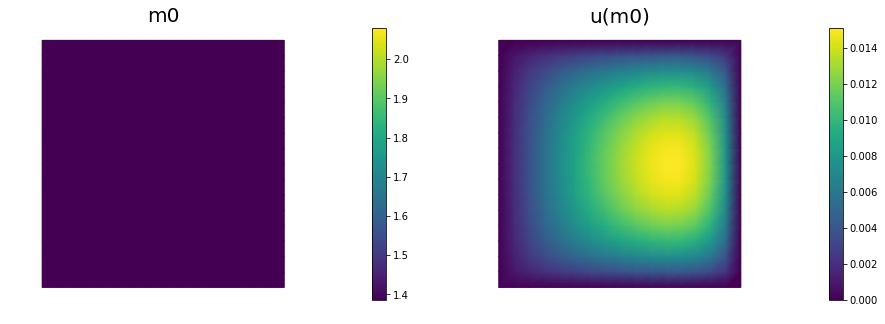
Steepest descent converged in 376 iterations
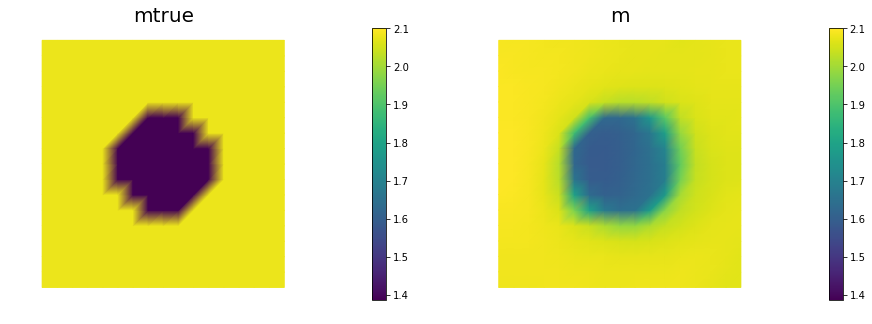
Copyright © 2016-2018, The University of Texas at Austin & University of California, Merced. All Rights reserved. See file COPYRIGHT for details.
This file is part of the hIPPYlib library. For more information and source code availability see https://hippylib.github.io.
hIPPYlib is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License (as published by the Free Software Foundation) version 2.0 dated June 1991.