Spectrum of the preconditioned Hessian misfit operator
The linear source inversion problem
We consider the following linear source inversion problem. Find the state and the source (parameter) that solves
Here:
-
is a finite dimensional vector that denotes noisy observations of the state in locations , . Specifically, , where are i.i.d. .
-
is the linear operator that evaluates the state at the observation locations , .
-
and are the parameters of the regularization penalizing the and norm of , respectively.
-
, , are given coefficients representing the diffusivity coefficient, the advective velocity and the reaction term, respectively.
-
, represents the subdomain of where we impose Dirichlet or Neumann boundary conditions, respectively.
1. Load modules
import matplotlib.pyplot as plt
%matplotlib inline
import dolfin as dl
import numpy as np
from hippylib import *
import logging
logging.getLogger('FFC').setLevel(logging.WARNING)
logging.getLogger('UFL').setLevel(logging.WARNING)
dl.set_log_active(False)
2. The linear source inversion problem
def A_varf(u,p):
return k*dl.inner(dl.nabla_grad(u), dl.nabla_grad(p))*dl.dx \
+ dl.inner(dl.nabla_grad(u), v*p)*dl.dx \
+ c*u*p*dl.dx
def f_varf(p):
return dl.Constant(0.)*p*dl.dx
def C_varf(m,p):
return -m*p*dl.dx
def R_varf(m_trial, m_test, gamma, delta):
return dl.Constant(delta)*m_trial*m_test*dl.dx \
+ dl.Constant(gamma)*dl.inner(dl.grad(m_trial), dl.grad(m_test))*dl.dx
def u_boundary(x, on_boundary):
return on_boundary and x[1] < dl.DOLFIN_EPS
class Hessian:
def __init__(self, A_solver, At_solver, C,B,R):
self.misfit_only = False
self.A_solver = A_solver
self.At_solver = At_solver
self.C = C
self.B = B
self.R = R
self.uhat = dl.Vector()
self.C.init_vector(self.uhat,0)
self.fwd_rhs = dl.Vector()
self.C.init_vector(self.fwd_rhs,0)
self.adj_rhs = dl.Vector()
self.C.init_vector(self.adj_rhs,0)
self.phat = dl.Vector()
self.C.init_vector(self.phat,0)
def init_vector(self, x, dim):
self.R.init_vector(x,dim)
def mult(self, x, y):
y.zero()
self.C.mult(x, self.fwd_rhs)
self.A_solver.solve(self.uhat, self.fwd_rhs)
self.B.transpmult(self.B*self.uhat, self.adj_rhs)
self.At_solver.solve(self.phat, self.adj_rhs)
self.C.transpmult(self.phat, y)
if not self.misfit_only:
y.axpy(1., self.R*x)
def solve(nx, ny, targets, gamma, delta, verbose=True):
rand_gen = Random()
mesh = dl.UnitSquareMesh(nx, ny)
Vh1 = dl.FunctionSpace(mesh, 'Lagrange', 1)
Vh = [Vh1, Vh1, Vh1]
if verbose:
print("Number of dofs: STATE={0}, PARAMETER={1}, ADJOINT={2}".format(
Vh[STATE].dim(), Vh[PARAMETER].dim(), Vh[ADJOINT].dim()) )
u_bdr = dl.Constant(0.0)
bc = dl.DirichletBC(Vh[STATE], u_bdr, u_boundary)
u_trial = dl.TrialFunction(Vh[STATE])
u_test = dl.TestFunction(Vh[STATE])
m_trial = dl.TrialFunction(Vh[PARAMETER])
m_test = dl.TestFunction(Vh[PARAMETER])
A,f = dl.assemble_system(A_varf(u_trial, u_test), f_varf(u_test), bcs=bc )
A_solver = dl.PETScLUSolver()
A_solver.set_operator(A)
At,dummy = dl.assemble_system(dl.adjoint( A_varf(u_trial, u_test) ), dl.Constant(0.)*u_test*dl.dx, bcs=bc )
At_solver = dl.PETScLUSolver()
At_solver.set_operator(At)
C = dl.assemble(C_varf(m_trial, u_test))
bc.zero(C)
R = dl.assemble(R_varf(m_trial, m_test, gamma, delta))
R_solver = dl.PETScLUSolver()
R_solver.set_operator(R)
mtrue = dl.interpolate(
dl.Expression('min(0.5,exp(-100*(pow(x[0]-0.35,2) + pow(x[1]-0.7,2))))',degree=5), Vh[PARAMETER]).vector()
m0 = dl.interpolate(dl.Constant(0.0), Vh[PARAMETER]).vector()
if verbose:
print( "Number of observation points: {0}".format(targets.shape[0]) )
B = assemblePointwiseObservation(Vh[STATE], targets)
#Generate synthetic observations
utrue = dl.Function(Vh[STATE]).vector()
A_solver.solve(utrue, -(C*mtrue) )
d = B*utrue
MAX = d.norm("linf")
rel_noise = 0.01
noise_std_dev = rel_noise * MAX
rand_gen.normal_perturb(noise_std_dev, d)
u = dl.Function(Vh[STATE]).vector()
m = m0.copy()
p = dl.Function(Vh[ADJOINT]).vector()
mg = dl.Function(Vh[PARAMETER]).vector()
rhs_adj = dl.Function(Vh[STATE]).vector()
# Solve forward:
A_solver.solve(u, -(C*m) )
# rhs for adjoint
B.transpmult(d-(B*u), rhs_adj)
# solve adj problem
At_solver.solve(p, rhs_adj)
#gradient
C.transpmult(p, mg)
mg.axpy(1., R*m)
H = Hessian(A_solver,At_solver,C,B,R)
solver = CGSolverSteihaug()
solver.set_operator(H)
solver.set_preconditioner( R_solver )
solver.parameters["print_level"] = -1
solver.parameters["rel_tolerance"] = 1e-9
solver.solve(m, -mg)
if solver.converged:
if verbose:
print ("CG converged in ", solver.iter, " iterations.")
else:
print( "CG did not converged.")
raise
# Solve forward:
A_solver.solve(u, -(C*m) )
if verbose:
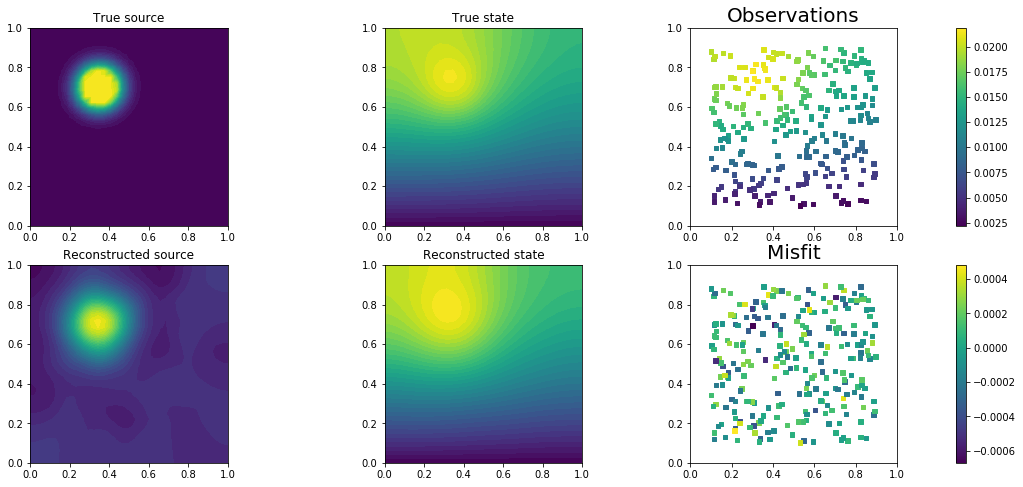
plt.figure(figsize=(18,8))
plt.subplot(2, 3, 1)
dl.plot(dl.Function(Vh[PARAMETER], mtrue), title = "True source")
plt.subplot(2, 3, 2)
dl.plot(dl.Function(Vh[STATE], utrue), title="True state")
plt.subplot(2, 3, 3)
nb.plot_pts(targets, d,mytitle="Observations")
plt.subplot(2, 3, 4)
dl.plot(dl.Function(Vh[PARAMETER], m), title = "Reconstructed source")
plt.subplot(2, 3, 5)
dl.plot(dl.Function(Vh[STATE], u), title="Reconstructed state")
plt.subplot(2, 3, 6)
nb.plot_pts(targets, B*u-d,mytitle="Misfit")
plt.show()
H.misfit_only = True
k_evec = 80
p_evec = 5
if verbose:
print ("Double Pass Algorithm. Requested eigenvectors: {0}; Oversampling {1}.".format(k_evec,p_evec))
Omega = MultiVector(m, k_evec+p_evec)
rand_gen.normal(1., Omega)
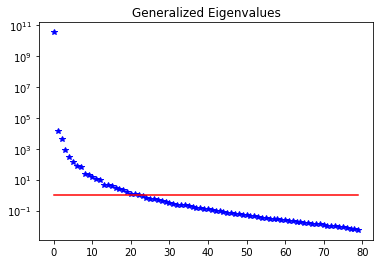
lmbda, U = doublePassG(H, R, R_solver, Omega, k_evec)
if verbose:
plt.figure()
nb.plot_eigenvalues(lmbda, mytitle="Generalized Eigenvalues")
nb.plot_eigenvectors(Vh[PARAMETER], U, mytitle="Eigenvectors", which=[0,1,2,5,10,15])
plt.show()
return lmbda, U, Vh[PARAMETER], solver.iter
3. Solution of the source inversion problem
ndim = 2
nx = 32
ny = 32
ntargets = 256
np.random.seed(seed=1)
targets = np.random.uniform(0.1,0.9, [ntargets, ndim] )
gamma = 1e-5
delta = 1e-9
k = dl.Constant(1.0)
v = dl.Constant((0.0, 0.0))
c = dl.Constant(0.1)
lmbda, U, Vm, nit = solve(nx,ny, targets, gamma, delta)
Number of dofs: STATE=1089, PARAMETER=1089, ADJOINT=1089
Number of observation points: 256
CG converged in 38 iterations.
Double Pass Algorithm. Requested eigenvectors: 80; Oversampling 5.
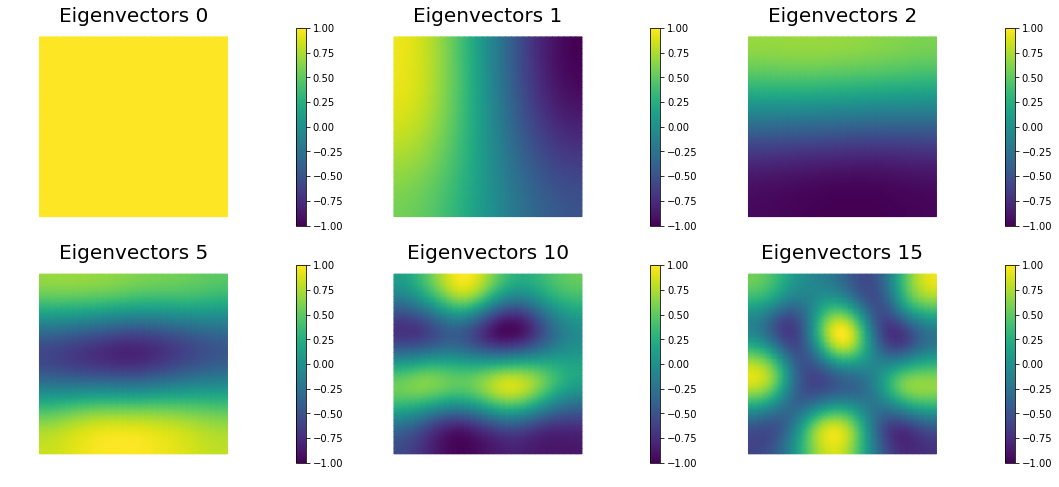
4. Mesh independence of the spectrum of the preconditioned Hessian misfit
gamma = 1e-5
delta = 1e-9
k = dl.Constant(1.0)
v = dl.Constant((0.0, 0.0))
c = dl.Constant(0.1)
n = [16,32,64]
lmbda1, U1, Vm1, niter1 = solve(n[0],n[0], targets, gamma, delta,verbose=False)
lmbda2, U2, Vm2, niter2 = solve(n[1],n[1], targets, gamma, delta,verbose=False)
lmbda3, U3, Vm3, niter3 = solve(n[2],n[2], targets, gamma, delta,verbose=False)
print ("Number of Iterations: ", niter1, niter2, niter3)
plt.figure(figsize=(18,4))
nb.plot_eigenvalues(lmbda1, mytitle="Eigenvalues Mesh {0} by {1}".format(n[0],n[0]), subplot_loc=131)
plt.ylim([1e-3,1e11])
nb.plot_eigenvalues(lmbda2, mytitle="Eigenvalues Mesh {0} by {1}".format(n[1],n[1]), subplot_loc=132)
plt.ylim([1e-3,1e11])
nb.plot_eigenvalues(lmbda3, mytitle="Eigenvalues Mesh {0} by {1}".format(n[2],n[2]), subplot_loc=133)
plt.ylim([1e-3,1e11])
nb.plot_eigenvectors(Vm1, U1, mytitle="Mesh {0} by {1} Eigenvector".format(n[0],n[0]), which=[0,1,5])
nb.plot_eigenvectors(Vm2, U2, mytitle="Mesh {0} by {1} Eigenvector".format(n[1],n[1]), which=[0,1,5])
nb.plot_eigenvectors(Vm3, U3, mytitle="Mesh {0} by {1} Eigenvector".format(n[2],n[2]), which=[0,1,5])
plt.show()
Number of Iterations: 37 38 36
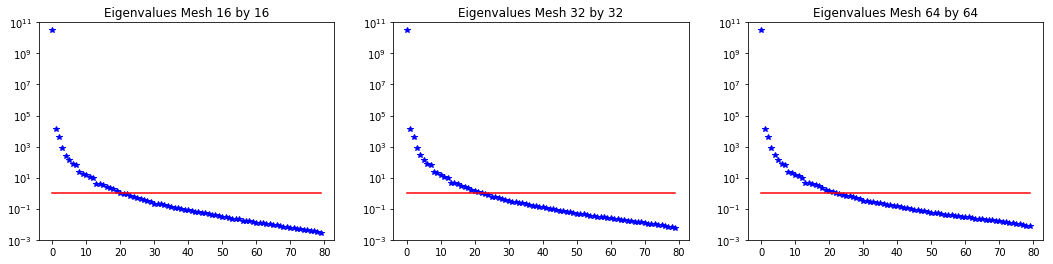
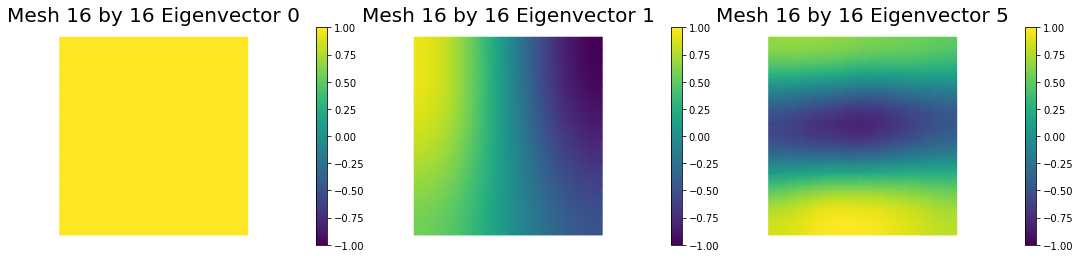
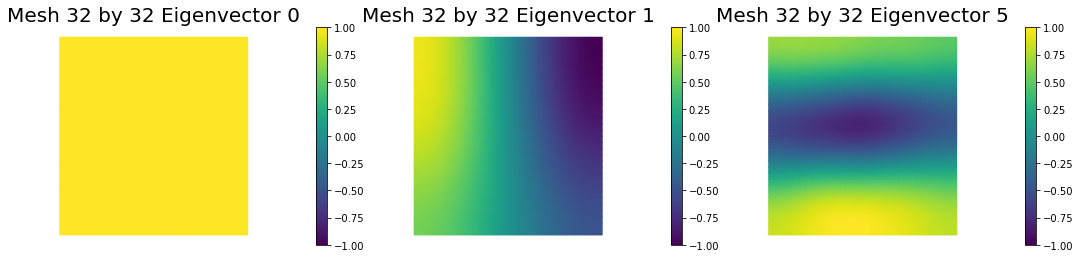
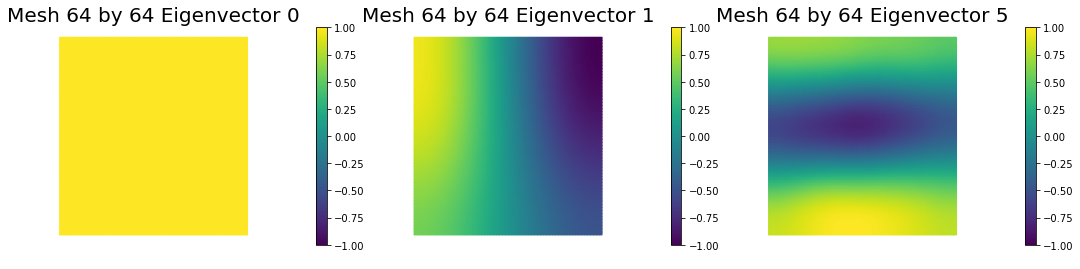
5. Dependence on regularization parameters
We solve the problem for different values of the regularization parameters.
gamma = [1e-4, 1e-5, 1e-6]
delta = [1e-8, 1e-9, 1e-10]
k = dl.Constant(1.0)
v = dl.Constant((0.0, 0.0))
c = dl.Constant(0.1)
lmbda1, U1, Vm1, niter1 = solve(nx,ny, targets, gamma[0], delta[0],verbose=False)
lmbda2, U2, Vm2, niter2 = solve(nx,ny, targets, gamma[1], delta[1],verbose=False)
lmbda3, U3, Vm3, niter3 = solve(nx,ny, targets, gamma[2], delta[2],verbose=False)
print ("Number of Iterations: ", niter1, niter2, niter3)
plt.figure(figsize=(18,4))
nb.plot_eigenvalues(lmbda1, mytitle="Eigenvalues gamma={0:1.1e}".format(gamma[0]), subplot_loc=131)
plt.ylim([1e-3,1e12])
nb.plot_eigenvalues(lmbda2, mytitle="Eigenvalues gamma={0:1.1e}".format(gamma[1]), subplot_loc=132)
plt.ylim([1e-3,1e12])
nb.plot_eigenvalues(lmbda3, mytitle="Eigenvalues gamma={0:1.1e}".format(gamma[2]), subplot_loc=133)
plt.ylim([1e-3,1e12])
nb.plot_eigenvectors(Vm1, U1, mytitle="gamma={0:1.1e} Eigenvector".format(gamma[0]), which=[0,1,5])
nb.plot_eigenvectors(Vm2, U2, mytitle="gamma={0:1.1e} Eigenvector".format(gamma[1]), which=[0,1,5])
nb.plot_eigenvectors(Vm3, U3, mytitle="gamma={0:1.1e} Eigenvector".format(gamma[2]), which=[0,1,5])
plt.show()
Number of Iterations: 23 38 64
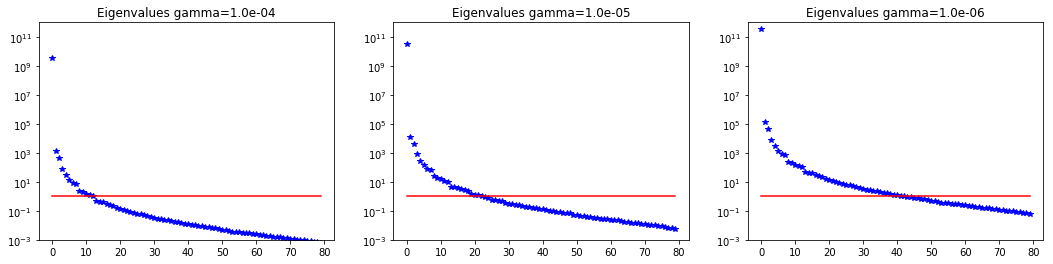
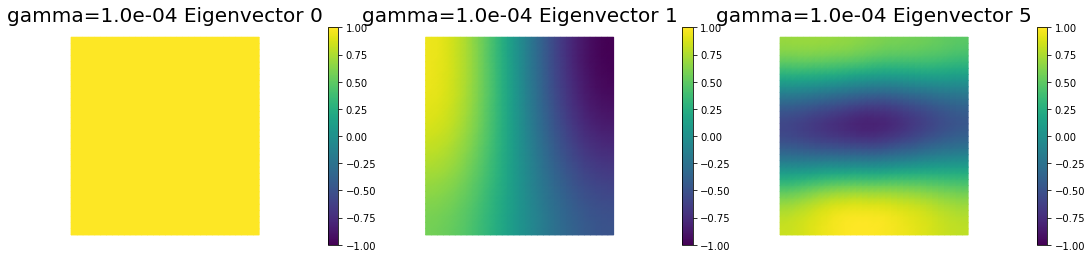
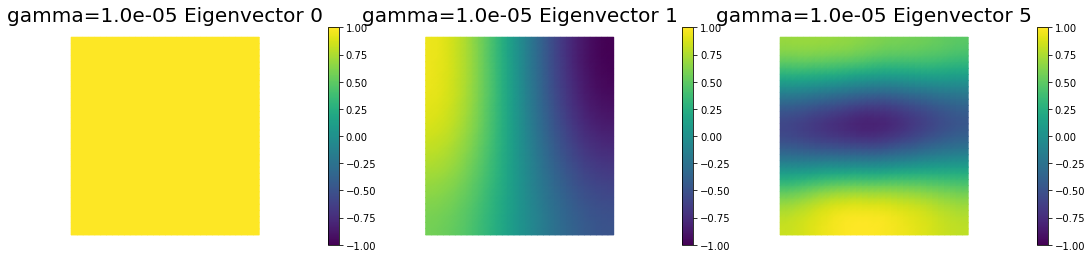
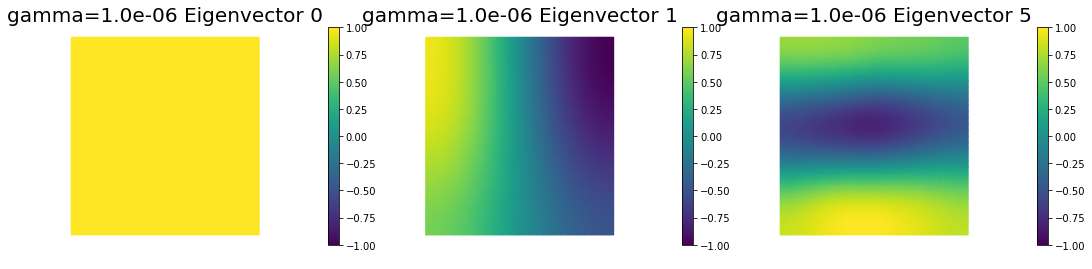
6. Dependence on the PDE coefficients
Assume a constant reaction term , and we consider different values for the diffusivity coefficient .
The smaller the value of the slower the decay in the spectrum.
gamma = 1e-5
delta = 1e-9
k = dl.Constant(1.0)
v = dl.Constant((0.0, 0.0))
c = dl.Constant(0.1)
lmbda1, U1, Vm1, niter1 = solve(nx,ny, targets, gamma, delta,verbose=False)
k = dl.Constant(0.1)
lmbda2, U2, Vm2, niter2 = solve(nx,ny, targets, gamma, delta,verbose=False)
k = dl.Constant(0.01)
lmbda3, U3, Vm3, niter3 = solve(nx,ny, targets, gamma, delta,verbose=False)
print ("Number of Iterations: ", niter1, niter2, niter3)
plt.figure(figsize=(18,4))
nb.plot_eigenvalues(lmbda1, mytitle="Eigenvalues k=1.0", subplot_loc=131)
plt.ylim([1e-2,1e14])
nb.plot_eigenvalues(lmbda2, mytitle="Eigenvalues k=0.1", subplot_loc=132)
plt.ylim([1e-2,1e14])
nb.plot_eigenvalues(lmbda3, mytitle="Eigenvalues k=0.01", subplot_loc=133)
plt.ylim([1e-2,1e14])
nb.plot_eigenvectors(Vm1, U1, mytitle="k=1. Eigenvector", which=[0,1,5])
nb.plot_eigenvectors(Vm2, U2, mytitle="k=0.1 Eigenvector", which=[0,1,5])
nb.plot_eigenvectors(Vm3, U3, mytitle="k=0.01 Eigenvector", which=[0,1,5])
plt.show()
Number of Iterations: 38 123 731
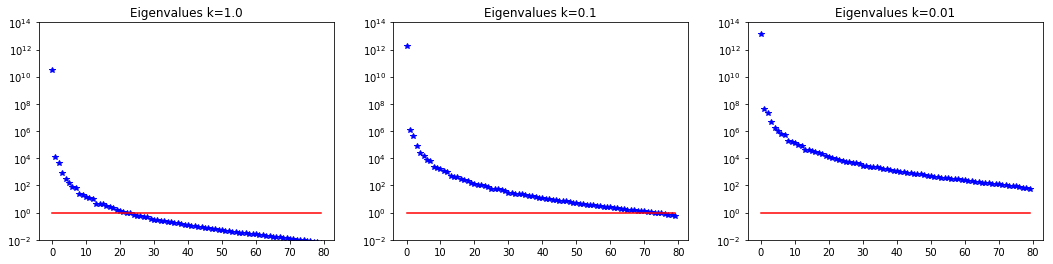
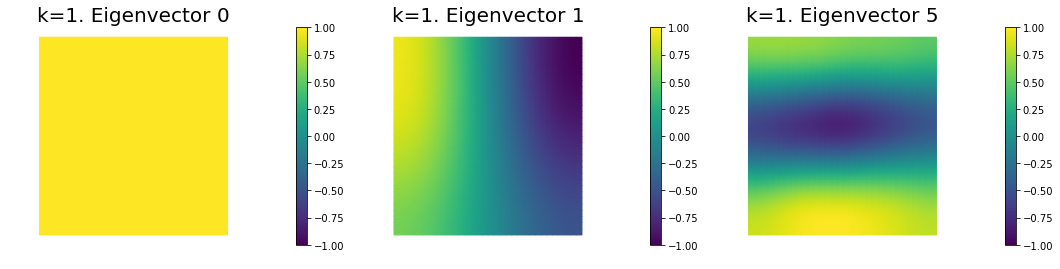
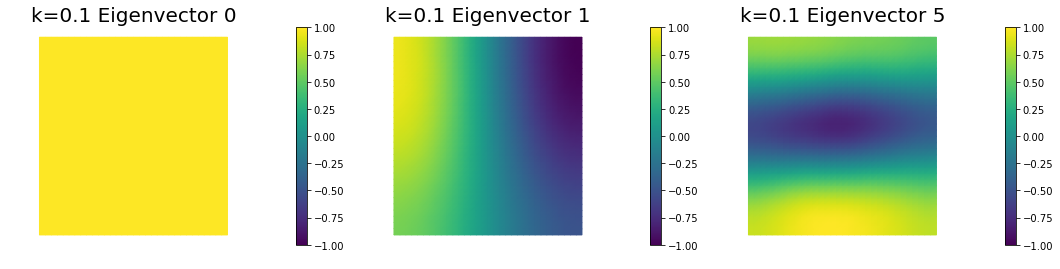
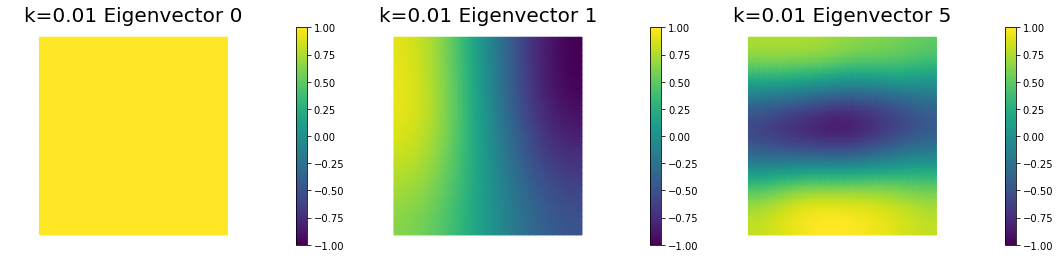
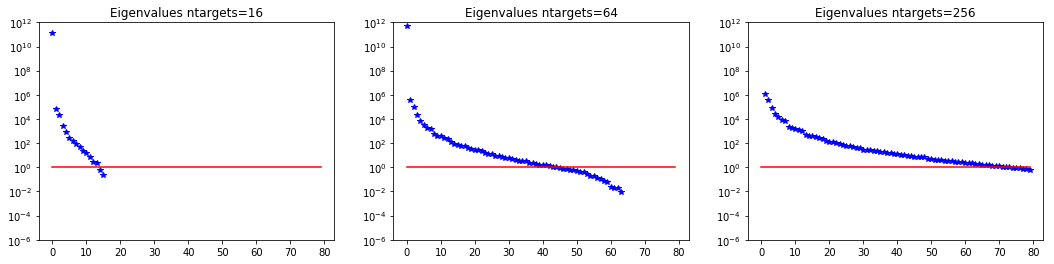
7. Dependence on the number of observations
ntargets = [16, 64, 256]
gamma = 1e-5
delta = 1e-9
k = dl.Constant(0.1)
v = dl.Constant((0.0, 0.0))
c = dl.Constant(0.1)
lmbda1, U1, Vm1, niter1 = solve(nx,ny, targets[0:ntargets[0],:], gamma, delta,verbose=False)
lmbda2, U2, Vm2, niter2 = solve(nx,ny, targets[0:ntargets[1],:], gamma, delta,verbose=False)
lmbda3, U3, Vm3, niter3 = solve(nx,ny, targets[0:ntargets[2],:], gamma, delta,verbose=False)
print ("Number of Iterations: ", niter1, niter2, niter3)
plt.figure(figsize=(18,4))
nb.plot_eigenvalues(lmbda1, mytitle="Eigenvalues ntargets={0}".format(ntargets[0]), subplot_loc=131)
plt.ylim([1e-6,1e12])
nb.plot_eigenvalues(lmbda2, mytitle="Eigenvalues ntargets={0}".format(ntargets[1]), subplot_loc=132)
plt.ylim([1e-6,1e12])
nb.plot_eigenvalues(lmbda3, mytitle="Eigenvalues ntargets={0}".format(ntargets[2]), subplot_loc=133)
plt.ylim([1e-6,1e12])
nb.plot_eigenvectors(Vm1, U1, mytitle="ntargets={0} Eigenvector".format(ntargets[0]), which=[0,1,5])
nb.plot_eigenvectors(Vm2, U2, mytitle="ntargets={0} Eigenvector".format(ntargets[1]), which=[0,1,5])
nb.plot_eigenvectors(Vm3, U3, mytitle="ntargets={0} Eigenvector".format(ntargets[2]), which=[0,1,5])
plt.show()
Number of Iterations: 31 91 123
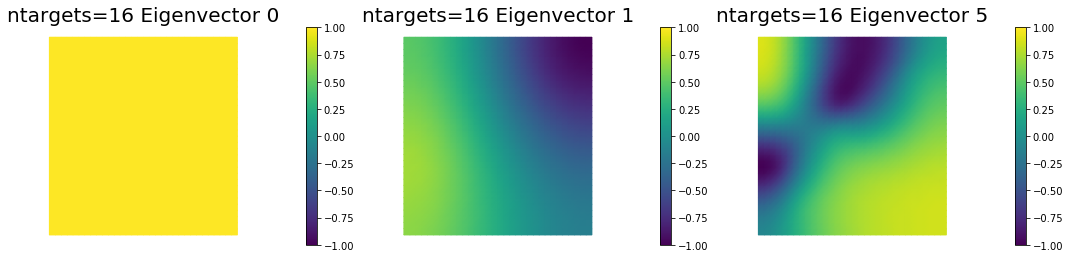
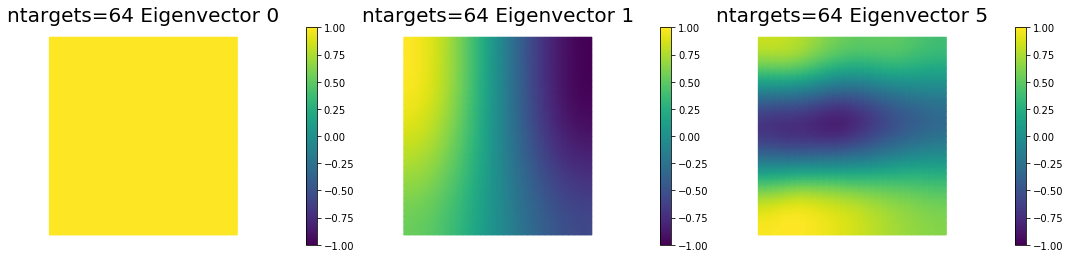
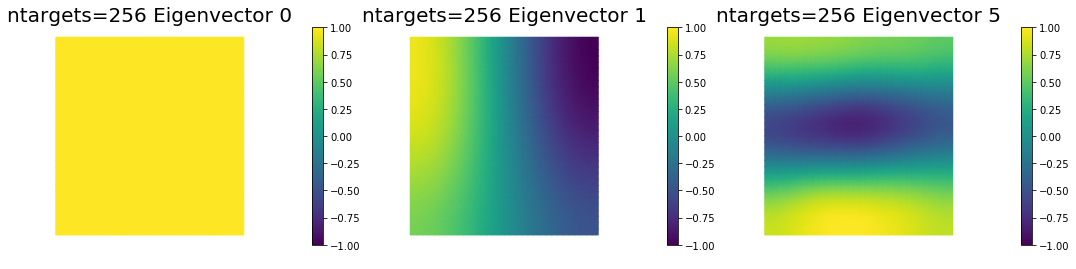
Copyright © 2016-2018, The University of Texas at Austin & University of California, Merced. All Rights reserved. See file COPYRIGHT for details.
This file is part of the hIPPYlib library. For more information and source code availability see https://hippylib.github.io.
hIPPYlib is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License (as published by the Free Software Foundation) version 2.0 dated June 1991.